Knickpoints and knickzones in river channels are areas of local steepening that can be caused by a number of factors such as lithologic differences or via transmission of changing erosion rates. A number of theories of channel incision suggest that if uplift rates changes, signals may propagate upstream through channel networks in the form of knickpoints and so locating knickpoints in river channels is often connected to interpreting the history of tectonic activity in an area through time.
Initial efforts to locate knickpoints and knickzones typically involved looking for waterfalls (a vertical knickpoint) or using channel long profiles. Later attempts integrated slope—area data because normalising for drainage area is essential if one is to compare headwaters with larger channels downstream.
Many authors report knickpoint locations but the methods used to identify them are frequently only semi-quantitatve (in that user input is required to directly select knickpoint location) and very difficult to reproduce. We have implemented a knickpoint detection algorithm that aims to minimise user inputs: it is scalable to user needs but uses advanced statistics to objectively describe and discriminate knickpoints. It is reproducible in that if the parameters of the method are reported and the same topographic data is used any author can reproduce the same map of knickpoint locations.
1. The methodology
The method has several steps, which will be outlined below. To summarise these steps here is a quick summary:
-
The method extracts channel steepness, \(k_{sn}\), using a statistical segmentation method of channel profiles based on Mudd et al., 2014 JGR. The knickpoint extraction does this automatically but detaiuls anbout the LSDTopoTools software for this can be found in our instructions for channel steepness extraction.
-
These segments of \(k_{sn}\) are derived using a Monte Carlo approach so there is noise at segment boundaries: the derived segments represent many iteration of the segmentation algorithm. To constrain the exact locations of the transitions a Total Variation Denoising (TVD) method from Condat (2013) is used to isolate the main \(k_{sn}\) variations.
-
We then use various algorithms for grouping and quantification of \(k_{sn}\) variations. You can read the detail in our paper.
-
Finally we implement analysis of the segmented elevation to isolate the knickpoints and knickzones using a variety of threhsolding and windowing methods.
We do not describe these methods in detail in this documentation: that information can be found in our upcoming ESURF paper and previous papers on our chi methods . These instructions are intended to simply get you using the software and extracting knickpoints and knickzones using your favourite topographic data.
2. Get the code
The knickpoint analysis tools lie within the chi mapping tool, so you can refer to the installation instructions of that tool. Once you install that you have the knickpoint extraction installed: you just need to switch on the correct flags in the parameter file.
Once you have compiled that tool (you can do that with either LSDTopoToolsSetup.py or by using make -f chi_mapping_tool.make you will then have the chi mapping tool program.
You use this program by calling a parameter file. These parameter files are explained in excruciating detail in other components of the LSDTopoTools documentation. These are instructions for the sorts of analyses you want done to your topographic data. They consist of keywords, which must be spelled correctly and are case sensitive, followed by a colon (:). The next section explains the keywords you need in your parameter file to run the knickpoint analysis.
By now you should have the chi_mapping_tools compiled and ready to run.
The next step is to create a parameter file to tell the code how to analyse your data.
3. The parameters
Here is a quick overview of how to set up and run the code. It will be much easier to understand if you have run a LSDTopoTools analysis before, so if that is not the case you can try the basic introductory analysis or try running a chi analysis first.
-
The beginning of your parameter file has information about the location and names of the input and ouput data: the software needs to know where your files are, what are they called and where to save them. The writing and reading filenames are just prefixes without extensions. If the read name is
Pollionnayit will readPollionnay.biland if it also is also the write fname it will write outputs likePollionnay_HS.bil. These lines has to be as follow:
read path: /home/boris/path/to/file/
read fname: name_of_dem_without_extension
write path: /home/boris/path/to/file/
write fname: prefix_for_output-
The next set of parameters controls the drainage network extraction (i.e. Basins and rivers), an exhaustive list is provided later. Here is a basic river extraction using a threshold of contributing pixels to initiate a channel:
threshold_contributing_pixels: 2000
minimum_basin_size_pixels: 200000
maximum_basin_size_pixels: 5000000
m_over_n: 0.45| If channel head extraction is important for your area (and if your DEM’s resolution allows it), guidance about extracting it can be found here. |
-
This methods highly depends on \(k_{sn}\), \(\theta\) and \(\chi\) extraction. A list of options for these chi-related ananlysis can be found in our sections on channel steepness analysis. As described in the manuscript it is important to constrain these in order to get relevant knickpoint magnitudes.
-
The options specific to the knickpoint analysis can be found in the this section of the parameters for the chi tool, however a few of these require some additional explanation:
-
The following parameters are linked to the \(k_{sn}\) extraction, using Mudd et al., 2014 JGR algorithm, they have been separated to the single \(k_{sn}\) extraction to avoid confusion when targetting these algorithm within LSDTopoTools:
-
force_skip_knickpoint_analysis: 2: Number of nodes skipped during each Monte Carlo iterations. 1 - 4. A lower value means a better fit to chi-plot profile, but also more sensitive to noise. -
force_n_iteration_knickpoint_analysis: 40: number of iteration for the segment testing. 20 is the minimum, over that value it does not change a lot the calculation. -
target_nodes: 80: Will set the number of nodes investigated per segment. combined withforce_skip_knickpoint_analysis, it will determine the size of the segments and thus the precision of the fitting (do you want the main trend or detail analysis?). Over 100 nodes, it becomes computationally expensive, over 120, you will need days of processing, over 150, probably months, over 200 you won’t be alive for the results. It therefore depends on your priorities. 50-100 nodes will fit most of the needs. -
Other parameters can be adjusted for the segment fitting algorithm, you can refer to the documentation or Mudd et al., 2014 JGR.
-
| Full description of how these parameters affect the extraction is described in the main manuscript and its supplementary materials. |
-
The following parameters directly control the knickpoint extraction:
-
ksn_knickpoint_analysis: true: switch on the analysis, as other analysis are available throughchi_mapping_tools.exe, namely Chi extraction, \(M_{\chi}\) extraction from Mudd et al., 2014 JGR and concavity index extraction from Mudd et al., 2018 Esurf. -
TVD_lambda: -1: define the regulation parameter for the Total Variation Denoising algorithm, adapted from Condat, 2013.-1will choose an automatic value depending on your concavity. This parameter depends on the magnitude of \(k_{sn}\) and can be adjusted manually. Higher \(\lambda\) values will produce a clearer signal but may inhibit or lower some knickpoint (\(\Delta k_{sn}\)). -1 is the default. The default values of this are (concavity then \(\lambda\) pairs): 0.2→0.5, 0.5→20, 0.7→300. So for example if you want fewer tightly clustered knickpoint you may use \(\lambda\) = 500 if your concavity is 0.5. The number of initial knickpoints is quite sensitive to this parameter, example of its effect is illustrated in the supplementary materials of the paper. -
kp_node_combining: 10: Determine the combining window for \(\Delta k_{sn}\) knickpoint. It avoids getting artifact knickpoints, but a high window can shift knickpoint location. A low window can scatter large knickpoints into successive small ones. 10 is the default. -
window_stepped_kp_detection: 100: Determine the window for windowed statistics on segmented elevation to detect stepped knickpoints. Low windows are unefficient to catch these steps. 100 is the default -
std_dev_coeff_stepped_kp: 8: Std deviation threshold to catch these variations. 7 - 9 gives a pretty reliable results. Lower value would produce many artifacts.
-
| Full description of how these parameters affect the extraction is described in the main manuscript and its supplementary materials. |
-
Please find bellow an example parameter file:
# Parameters for selecting channels and basins
threshold_contributing_pixels: 2500
minimum_basin_size_pixels: 200000
maximim_basin_size_pixels: 500000
test_drainage_boundaries: false
# Parameters for chi analysis
A_0: 1
m_over_n: 0.35
n_iterations: 20
target_nodes: 80
minimum_segment_length: 10
sigma: 10.0
skip: 2
# ksn_knickpoint_analysis
ksn_knickpoint_analysis: true
TVD_lambda: -1
std_dev_coeff_stepped_kp: 84. Running the code and outputs
-
Once you have set up the chi mapping tool you simply run the code pointing to your parameter file:
$ chi_mapping_tool.exe /path/to/parameter/file parameter_file.driver -
Your life will be easier if you ensure the paths and filenames are all correct. Getting these wrong is the most common reason the program fails. The 2nd most common reason the program fails is that the DEM is too large: LSDTopoTools requires substantial amounts of memory and large DEMs will result in something called segmentation faults.
-
The analysis will then churn away for some time, printing all manner of things to screen which hopefully are not error reports and that you can safely ignore.
-
At the end of the process, you will have several new files:
-
_ksnkp.csv: This has summary information on the detected knickpoints (e.g., lat,long,X,Y,elevation,ksn…). -
_ksnkp_mchi.csv: This is a large file with information about all the segments, as well information about \(k_{sn}\) values, chi-elevation plots, and loads more information. It is the workhorse file for the plotting routines. -
_ksnkp_raw.csv: Similar to the_ksnkp.csvfile but contains all unfiltered knickpoint locations so that users can develop their own extraction algorithms. -
_ksnkp_SK.csv: This files contains basic information about each source keys, its length for instance, and speed up the plotting routines when selecting specific channel length.
-
5. Plotting
Running the code will produce a number of files with data that can be visualised with either geographic information software or our own plotting routines within our LSDTopoTools python visualisation toolchain. This section details the automated plotting routines we have provided in support of the knickpoint algorithms. These have been used to generate the figures in the manuscript and its supplementary materials. It also provide basic tools to (i) select which river or basin you want to plot (ii) plot statistical description of the knickpoint magnitude and (iii) apply thresholds to isolate main entities in a reproducible way.
5.1. The philosophy
We provide automated and open-source plotting option using Python code and pakages. Many plotting option are automated thanks to the formatted outputs from the c++ code and does not require coding to be used. Plotting routines are accessible via command line and propose a range of customization. For advanced plotting, we recommend to code directly in Python as Matplotlib package produce amazing publication-ready figures.
5.2. How to plot
Our plotting tools rely on the LSDTopoTools python toolchain. You can read about how to set that up in our visualisation section.
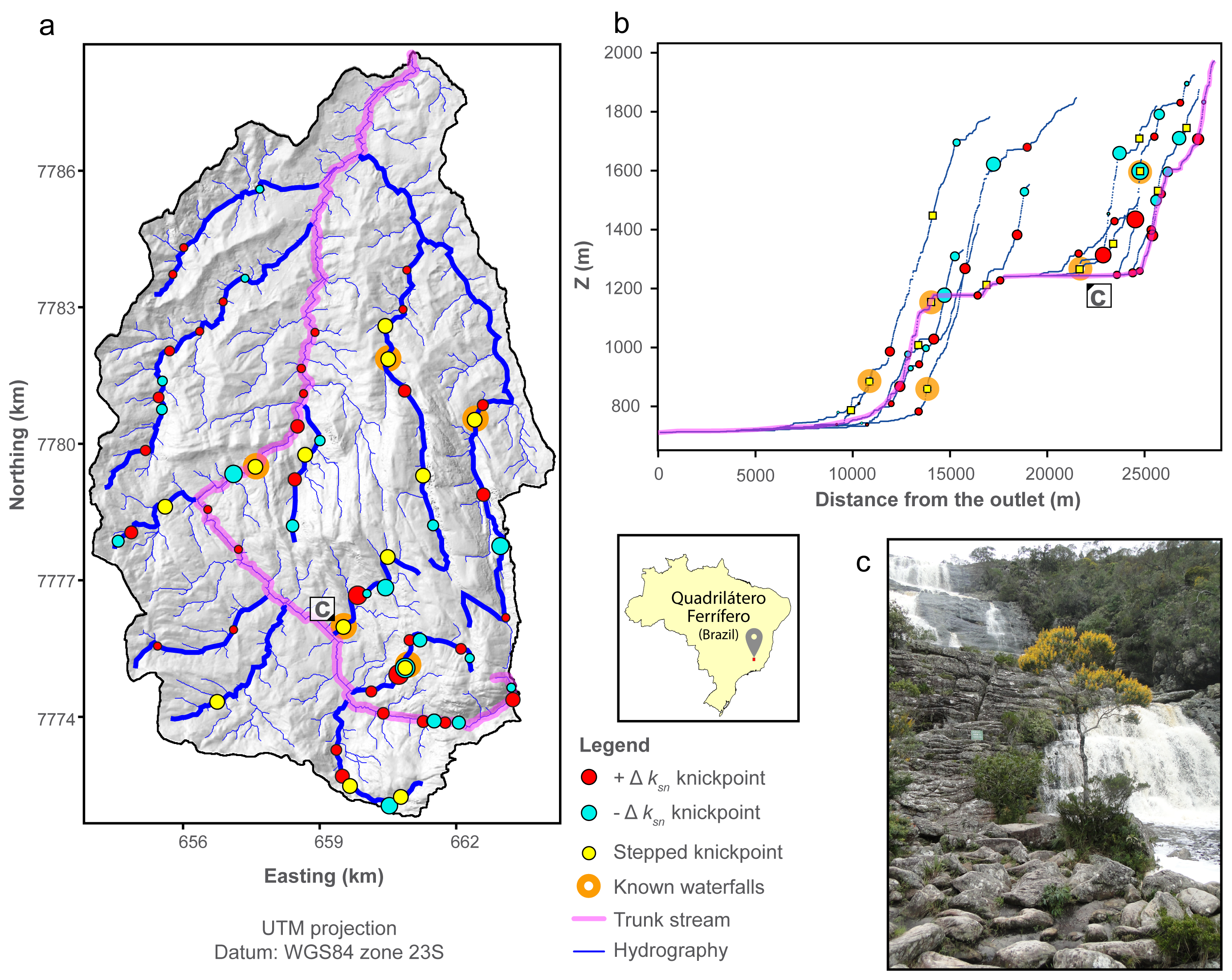
The knickpoint algorithm produces large datasets. We provide available plotting routines to help the user dealing with this. These routines are useful (at least we think so) for giving users a first overview of the data, to descirbe its statistics, to thin the knickpoint and knickzone dataset, and produce publication-ready plots thanks to the matplotlib python package. All the routines are implemented in the LSDMappingTools suite. We will describe these plotting routines with an applied example from the paper: The Smugglers basin (Santa Cruz Island, USA).
Here is a guide on how to use the automated plotting routines:
-
In a terminal, you need first to navigate to the LSDMappingTools folder, depending on your OS, you need to use the equivalent of
cd. example on linux:cd /home/boris/Desktop/LSDMappingTools/, or on Windows:cd C:\windows\something\LSDMappingTools\ -
Right now we distribute our python toolchain as a conda environment so you will need to activate it (using the command
activate LSDTT). -
All the plotting routine have a common start: you need to call the python script and flag the path and common prefix of all the files as follow:
python PlotKnickpointAnalysis -dir /home/boris/Desktop/GIS/massif_central/ -fname massif_central_DEM -
The second step is optional: you can select specific river to plot:
-basin_keys 1,5will only select the basin 1 and 5;-source_keys 54,0,12,18will only select rivers 54,0,12 and 18 and finally-min_sl 10000will only select rivers with a flow length greater than 10000 metres. None of these flags means all channels will be selected. -
You now have the specific rivers you want to analyse: you then may need to sort out your main knickpoints to thin the dataset and only keep the relevant ones.
-statplot Truewill plot histogram distributions of stepped knickpoints and \(\Delta k_{sn}\). It provides a first order statistical description to sort knickpoints. It can be customized with-nbh 80to force a number of bins. -
-ksnPs Trueis a calibrating plotting option that plots a \(\chi\) - \(k_{sn}\) profile with raw and denoised \(k_{sn}\) values in order to check the effect of the \(\lambda\) parameter, which controls the degree of smoothing using the TVD algorithm. This is described in the supplementary materials of the main manuscript. -
-cov 3,5,2is a parameter that apply a basic cut off value on the knickpoints, in order to select the main ones. In this example it will select knickpoint with \(\Delta k_{sn}\)< -3, \(\Delta k_{sn}\) > 5 and \(\Delta z_{seg}\) > 2. -
-GS 1,1.5,2,5controls the RELATIVE sizing of your knickpoints for each type: in this example, all the \(\Delta k_{sn}\) knickpoints with a magnitude < 1 will have the same minimum size ; all the step knickpoints with a magnitude < 1.5 will have the minimum size ; all the \(\Delta k_{sn}\) knickpoints with a magnitude > 2 will have the same maximum size and all the step knickpoint with a magnitude >5 will have the same maximum size. In between these range, the size evolve lineraly. -
-coeff_size_kp 30will set the global size of the knickpoints markers: larger number = bigger markers. It doesn’t manipulate data but only modifies the appearance of the plots. -
Finally you need to tell the plotting routine which families of plots you need (we have plachaged these up so the script produces loads of plots for you):
-clas Truewill produce basin-wide profiles and map of knickpoints whereas-clasriv Truewill produce plots for each river. -
Done! Here is a full command line to copy-paste and adapt:
-
Calibration:
python PlotKnickpointAnalysis -dir /home/boris/Desktop/GIS/massif_central/ -fname massif_central_DEM -basin_keys 1,5 -ksnPs True -statplot True -nbh 75 -
Plotting:
python PlotKnickpointAnalysis -dir /home/boris/Desktop/GIS/massif_central/ -fname massif_central_DEM -basin_keys 1,5 -cov 3,5,2 -GS 1,1.5,2,5 -coeff_size_kp 30 -clas True
-
5.3. Plotting with other tools (e.g. GIS, ArcGIS or you own code in R, Python, or other programming languages)
The data outputs are designed to be compatible with common geographic information systems. The knickpoint method produces a set of raster and csv files that one can import and plot with your own plotting routines. X being the prefix of all the output, the algorithm generates additional rasters: hillshaded DEM X_hs.bil and delimination of analysed basins X_AllBasins.bil.
It also generates the point-based data. X_ksnkp_mchi.csv contains all the river-related data:
| Column | Data |
|---|---|
X |
Easting in UTM (Same zone than the Raster). |
Y |
Northing in UTM (Same zone than the Raster). |
Longitude |
Longitude in WGS84. |
Latitude |
Latitude in WGS84. |
chi |
The chi coordinate. |
elevation |
Elevation of the river point in meters. |
flow_distance |
Flowing distance from the outlet in meters. |
drainage_area |
Drainage area of the point in meter square. |
m_chi |
Chi-elevation steepness calculated with Mudd et al., 2014 JGR algorithm. Is k_sn if A0 = 1. |
source_key |
Unique ID of this river, from source to the point where it reaches a longer river or the outlet of the basin. |
basin_key |
Unique basin ID. |
Other Columns |
Other columns are for debugguing purposes, Ignore them, I’ll remove them after the publication of the paper. |
X_ksnkp.csv contains all the knickpoint-related data:
| Column | Data |
|---|---|
X |
Easting in UTM (Same zone than the Raster). |
Y |
Northing in UTM (Same zone than the Raster). |
Longitude |
Longitude in WGS84. |
Longitude |
Longitude in WGS84 |
Latitude |
Latitude in WGS84. |
chi |
The chi coordinate. |
elevation |
Elevation of the river point in meters. |
flow_distance |
Flowing distance from the outlet in meters. |
drainage_area |
Drainage area of the point in meter square. |
delta_ksn |
Magnitude of the knickpoint "slope-break" component quantified by it’s drop/increase of ksn. |
delta_segelev |
Magnitude of the knickpoint "vertical-step" component quantified by sharp increase of segmented elevation. |
sharpness |
Cumulated Chi distance between all the combined knickpoints. 0 for single knickpoint. |
sign |
1 if increase of delta_ksn, -1 if decrease. |
source_key |
Unique ID of this river, from source to the point where it reaches a longer river or the outlet of the basin. |
basin_key |
Unique basin ID. |
Other Columns |
Other columns are for debugguing purposes, Ignore them, I’ll remove them after the publication of the paper, or I will forget. I will probably forget. So ignore it anyway. |
The two other files are to help the automated plotting routines to be faster.